Contributing Data to the Human Cell Atlas
Overview
The following instructions are for scientists wanting to contribute datasets to the Human Cell Atlas (HCA). For data to be included in atlases, data contributors must provide the following data and metadata. The data and metadata are stored in three platforms to ensure adequate protection is provided to sensitive information (e.g. FASTQ and Tier 2 metadata) and non-personal data is more openly available. For the list of metadata fields, click here.
Data Platforms
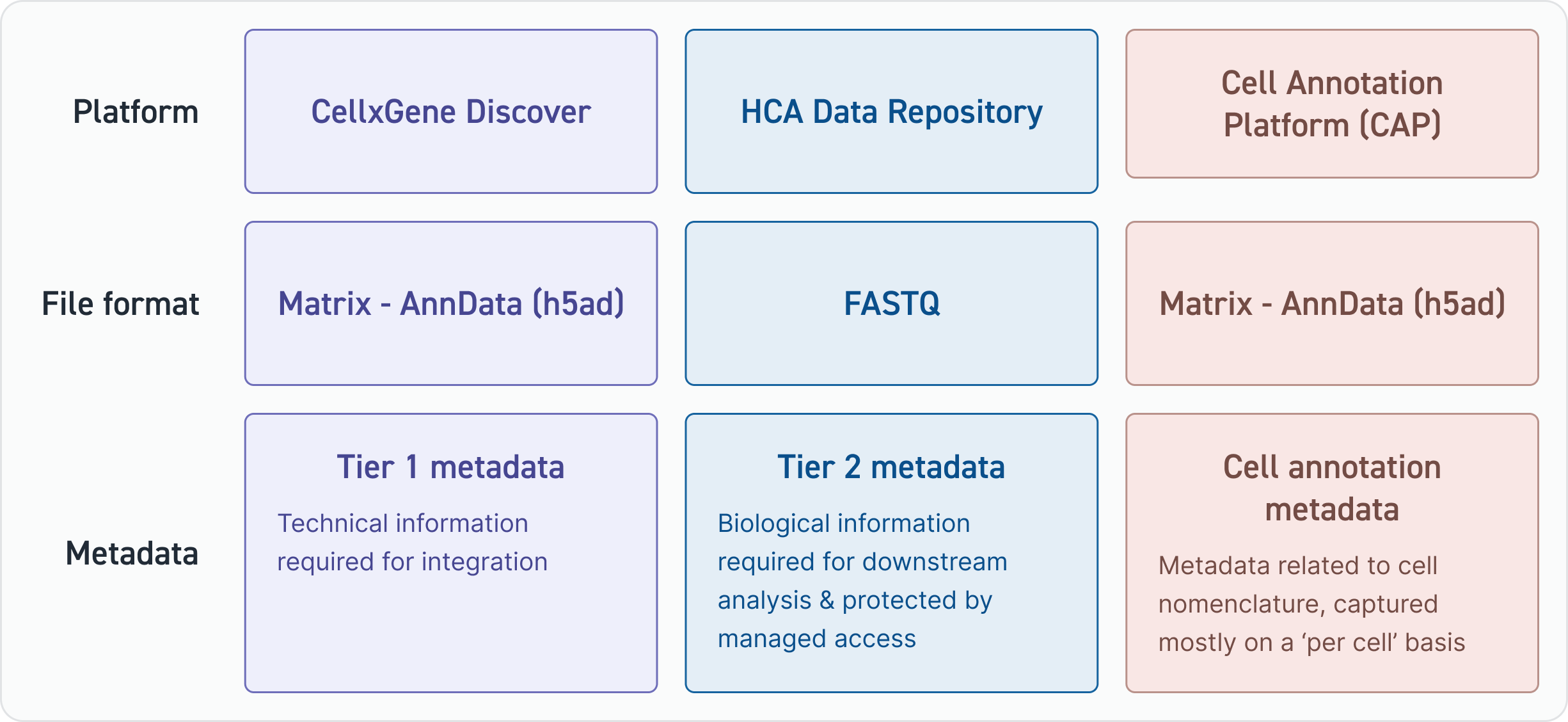
The CellxGene Discover platform stores matrices (in AnnData format) and Tier 1 metadata which are the core building blocks of atlases. Tier 1 metadata fields are typically ‘technical’ in nature (e.g. whether there was cell enrichment or depletion). For support creating an AnnData file, refer to this tutorial.
The HCA Data Repository stores FASTQ files and the Tier 2 metadata which may contain personal information. To provide sufficient protection to this information, the HCA Data Repository operates a managed access service through the DUOS platform. Access to these datasets and metadata requires the submission of a Data Access Request which is reviewed by HCA’s global Data Access Committee.
The Cell Annotation Platform stores matrices and cell annotation metadata in a specially designed portal with features that allow members of the HCA community to jointly annotate data.
Inclusion Criteria
HCA’s Biological Networks (open networks of scientists responsible for constructing atlases) are responsible for choosing the datasets that end up in HCA atlases. Not all datasets submitted to the HCA Data Portal will ultimately form part of an atlas.
Contributing Unpublished Data
HCA welcomes the contribution of unpublished data to atlases
HCA welcomes the contribution of unpublished data to atlases and has put in place mechanisms to protect the data prior to the publication of atlases. Importantly, contributors of unpublished data are encouraged to make their data publicly available immediately via the HCA Data Repository, in accordance with the HCA Data Release Policy.
If the data contributors are unable or decline to do so, they must, at minimum, agree to make their data publicly available via the HCA Data Repository (for raw sequence data and detailed Tier 2 metadata) and Chan Zuckerberg CELLxGENE (for gene expression matrices, Tier 1 metadata, and broad demographic metadata) as soon as the Atlas that incorporates it is published in a peer-reviewed scientific journal.
Embargoing unpublished data
If you are contributing unpublished data, you may request that your data be “embargoed” in a private HCA data repository until the organ/tissue/system atlas you have contributed to is published. At that point, your data will be made available publicly via the HCA Data Explorer and linked to the corresponding Atlas page on the HCA Data Portal. Data contributors will be notified before their data is made public. Access to embargoed data will be restricted to HCA Data Wranglers, the corresponding Atlas integration team, and anyone else specified by the data contributor.
HCA’s Unpublished Data Policy
For more information, refer to HCA’s Unpublished Data Policy.
Protecting Managed Access Data
HCA has a managed access service. Data submitted to the HCA Data Repository can be protected by managed access if elected to by the data contributor or by HCA. Access to managed access data is controlled through the HCA Data Access Committee. Scientists seeking access to these data must submit a data access request which is reviewed by the HCA Data Access Committee.
Submitting Metadata to Other Repositories
Data contributors are asked to notify HCA if biological metadata is submitted to other data repositories.